TEMPERATURE DEPENDENCE OF THE CYCLIZATION OF GUANINE AND CYTOSINE MIX HEXANUCLEOTIDES WITH WATER-SOLUBLE CARBODIIMIDE AT 0 - 75 °C
Kunio Kawamura*, Noriyuki Nakahara, Fumitaka Okamoto, and Noriko Okuda
Department of Applied Chemistry, Graduate School of Engineering, Osaka Prefecture University,
Sakai, Osaka 599-8531, Japan
Fax: 0722-54-9903; Email: kawamura@chem.osakafu-u.ac.jp
(Received 4 September 2003 Accepted 10 October 2003)
(Abstract)
The temperature dependence of pseudo-first-order rate constants (kcyc) of the cyclization of hexanucleotides 5'-d(pGCGCG)rC and 5'-d(pGCCCG)rG was investigated in the presence of 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDAC) and imidazole at 0 - 75 °C. Although the association between the elongating oligomer and the activated monomer is necessary for the success of the formation of oligonucleotides on a polynucleotide template or a clay catalyst, the phosphodiester bond formation in the case of cyclization readily occurs without these catalysts. Thus, the cyclization is regarded as a simple model of the formation of phosphodiester bond. The rate constants (kcyc) were compared with those of the cleavage of the ribose phosphodiester bond, in which the cyclization was more than 50 times faster than the cleavage at 75 °C. On the basis of this fact, it is deduced that the prebiotic formations of RNA oligomers such as the template-directed reaction and the mineral catalyzed oligomerization would be possible at high temperatures if the association between elongating oligomer and activated monomer (or activated oligomer) is sufficiently strong.(Keyword) Cyclization of oligonucleotide, Temperature dependence of the formation and degradation of phosphodiester bond, Hydrothermal origin of life, Oligonucleotide, RNA, Phosphodiester bond formation, Hydrolysis of phosphodiester bond, Cyclicnucleotide, RNA world
Introduction
The discovery of catalytic functions of ribonucleic acids (RNA) suggests that RNA or RNA-like molecules played central roles for the emergence of genetic information on the primitive earth environments (RNA world hypothesis) [1]. If the RNA world hypothesis is correct then RNA could have been accumulated under the prebiotic earth conditions. The condensation reactions of nucleotides with and without using activated nucleotide monomer have been investigated as primitive polymerase models of oligonucleotides. Examples include the condensation of activated nucleotide monomers on a polynucleotide template [2,3], and the spontaneous oligomerization of activated nucleotide monomer in the presence of several catalysts [4,5]. In addition, the condensation reactions of oligonucleotides using 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDAC) were also investigated [6-9]. In our group, the condensation reaction of hexanucleotides was studied in the presence of EDAC at 0 °C, in which the cyclization of hexanucleotide proceeds efficiently [10].On the other hand, it is widely believed that hydrothermal systems such as hydrothermal vents in the deep ocean played important roles for the emergence of life (the hydrothermal origin of life hypothesis) [11]. Thus, the RNA world hypothesis seems to be inconsistent with the hydrothermal origin of life hypothesis since it is imaginable that RNA could not have possessed sufficient stability for preserving information and catalytic ability under the hydrothermal conditions [12]. Although both the rates of the formation and decomposition of RNA should determine the accumulation of RNA molecules under the primitive earth conditions, there have been few investigations on the temperature dependence of the prebiotic formation and decomposition. The comparison of the rates of the formation and degradation of oligonucleotides is essential to estimate the accumulation behavior of RNA. Thus, we have been carried out comparative kinetics of the formation and degradation of RNA from the viewpoint of the hydrothermal origin of life [13,14].
Our previous study on kinetics of the template-directed reaction (TD reaction) at 40 - 80 °C elucidated that the rate constants of the formation of 3-mer and longer are greater than those of the degradation of oligonucleotides formed by the TD reaction even at 80 °C and higher [13]. However, the actual yield of the TD reaction was very low at 80 °C. The reason is due to that the relative rate of the formation of 2-mer notably decreases at 80 °C and higher. Thus, it was concluded that the TD reaction is not efficient at over 80 °C unless the formation of 2-mer is enhanced. Fortunately, the comparison between the rate constants of the formation of oligoguanylate (oligo(G)) by the TD reaction and the hydrolysis of oligo(G) at elevated temperatures was succeeded in the TD reaction system, since both the formation and degradation of oligo(G) obeyed pseudo-second order processes.
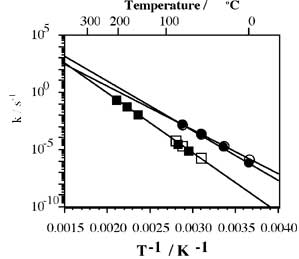
Recently, we found that the cyclization of hexanucleotides readily occurs in the presence of water-soluble carbodiimide (Fig. 1), which is highly efficient compared with the dimerization of activated nucleotide oligomers in the absence of the polynucleotide template or the clay catalyst [10]. In addition, since the cyclization reaction obeys a pseudo-first-order process, the direct comparison of the rate constants of the formation and cleavage of phosphodiester bond is possible. These facts indicate that the cyclization of oligonucleotides is a suitable model reaction to evaluate the rate constants of the phosphodiester bond formation at high temperatures. In the present study, the scope of the cyclization reactions of hexanucleotides 5'-d(pGCGCG)rC (oligo-6a) and 5'-d(pGCCCG)rG (oligo-6b) was investigated, in which the rate constants (kcyc) were determined at 25 - 75 °C. In addition, the rate constants of the cleavage of the ribose phosphodiester bond of dCrCdGdG (kdeg) were determined in the presence of EDAC at elevated temperatures. The thermodynamic parameters of the cyclization and cleavage of the phosphodiester bond were also determined from the temperature dependence of these rate constants. Possibility of the accumulation of RNA is discussed on the basis of the comparison of kcyc and kdeg.
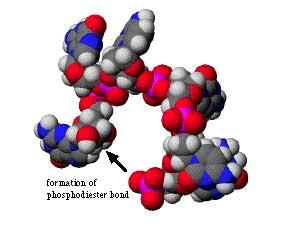
Figure 1. A perspective view of the formation of phosphodiester bond in hexanucleotide with van der Waals radii. Hexanucleotide is oligo-6b, in which the activated terminal is not shown.
Experimental
Oligonucleotides oligo-6a, oligo-6b, dCrCdGdG, d(pC5)rC, d(pC6)rC, and d(pC7)rC were purchased from GENSET (France) as HPLC purified grade. All other reagents used were of analytical grade.The condensation reactions were performed in an aqueous solution containing 0.2 M NaCl, 0.075 M MgCl2, 0.1 M imidazole, 0.2 M EDAC, and 0.05 - 0.1 mM oligo-6a, oligo-6b or oligocytidylic acid (oligo(C)) at 25 - 75 °C. The solution containing EDAC was freshly prepared for each reaction. The pH of the sample solutions was adjusted with 0.1 M NaOH or 0.1 M HCl solution. The mixture was allowed to stand for 24 h at 25 °C, 3 h at 50 °C, and 30 min at 75 °C. Aliquot of sample was periodically withdrawn and immediately quenched in liquid nitrogen to stop the reaction. Samples were analyzed by HPLC on a DNA-NPR anion exchange column from TOSOH Co., Tokyo, Japan using a gradient of 0 - 1.2 M NaCl at pH 11 with 0.02 M 2-amino-2-hydroxymethyl-1,3-propanediol.
The degradation of dCrCdGdG was investigated in a buffer solution, which is the same as used for the condensation reactions of oligo-6a and oligo-6b. Products were analyzed by the reversed-phase HPLC on a ODS-2 column from GL Science Co., Tokyo Japan using a gradient of 0.02 M NaH2PO4, 0.005 M tetrabuthylammonium bromide (TBABr) in water at pH 3.5 mixed with 0.02 M NaH2PO4, 0.005 M TBABr in 60% CH3OH at pH 3.5.
The melting temperatures (Tm) of d(GCGCGC) and d(GGCCCG) were determined in aqueous solutions containing 0.2 M NaCl, 0.075 M MgCl2, 0.1 M imidazole, and 0.67 mM oligo-6a or oligo-6b at pH = 8.0 using a CSC 6100 Nano II differential scanning calorimeter (DSC) (Calorimetry Sciences Corp., USA).
Results and Discussion
Stability of EDACThe stability of EDAC was investigated on the basis of our previous study [10]. UV spectra of the solution containing NaCl, MgCl2, imidazole, and EDAC was monitored at 200 - 300 nm at 0 - 75 °C, where the maximum wavelength was 234 nm [10]. The result showed that absorbance at 234 nm decreased to 85 % of the initial amount of EDAC after 7 days at 0 °C, 87 % after 16 h at 25 °C, 77 % after 2 h at 50 °C. The absorbance at 234 nm slightly increased after 30 min at 75 °C. The reaction curves at the beginning of the cyclization reactions were used for the determination of the rate constants of the disappearance of hexanucleotides, at which the influence of the degradation of EDAC was possible to be ignored.
Reaction behavior of cyclization at 0 - 75 °C
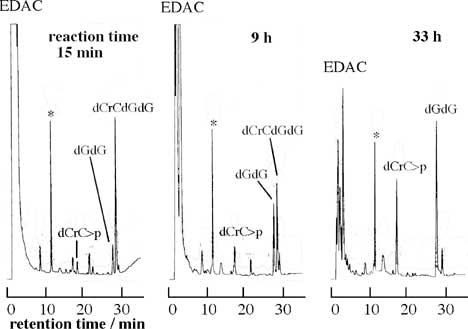
The condensation reaction of oligo-6a and that of oligo-6b were monitored by HPLC and two main peaks appeared (peak-1 and peak-2), where peak-1 was assigned to oligo-6a or oligo-6b [10]. The characterization of peak-2 was carried out in our previous study using specific enzymatic hydrolyses; the peak-2 fraction was assigned to cyclic hexanucleotide (cyclic-oligo-6) [10]. It was confirmed that peak-2 disappeared by the treatment with ribonuclease T2 (RNaseT2) and was not changed by the alkaline phosphatase treatment. The reaction curves at 25 - 75 °C are shown in Fig. 2. A relatively large amount of the dimerized product (12-mer) of oligo-6b was detected in the condensation of oligo-6b at 0 °C, and a very small amount of the dimerized product of oligo-6a was barely detected. The yield of the dimerized product of oligo-6b became negligible at 25 °C and higher temperatures. Since imidazole was necessary for the cyclization reactions of oligo-6a and oligo-6b, the formation of imidazolide onto the 5'-terminal phosphate of hexanucleotide is assumed in the presence of EDAC. The formation of imidazolide is supported by several studies [6-8,10]. Cyclic hexanucleotides were rapidly formed from oligo-6a and oligo-6b, but the decomposition of cyclic-oligo-6 was hardly observed (Fig. 2). Thus, the cyclization is obviously faster than the degradation of cyclic-oligo-6. The reaction for the case of oligo-6a is expressed in equations (1) and (2).
5'-d(pGCGCG)rC → cyclic-oligo-6a (1)
cyclic-oligo-6a → 5'-d(GCGCG)rC>p (2)
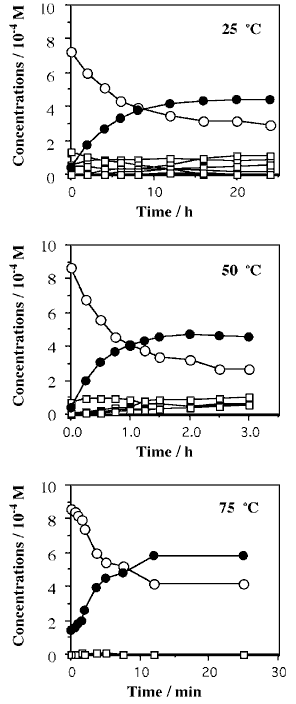
Pseudo-first-order rate plots of the disappearance of oligo-6a and oligo-6b showed that the reactions obey pseudo-first-order kinetics at the beginning of the reactions, at which the reactions proceed about 50 % (Fig. 2). The reaction was gradually slowed from the pseudo-first-order process. This is mainly caused by the degradation of EDAC and possibly by the degradation of cyclic-oligo-6, so that the rate constants were determined from the linear part of the pseudo-first-order rate plots (kcyc) (Fig. 3, Table 1). The magnitudes of kcyc are not much different between oligo-6a and oligo-6b.
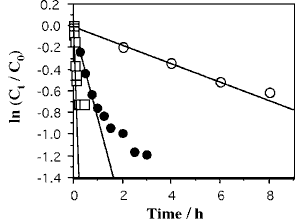
Figure 3. Pseudo-first-order rate plots for the disappearance of oligo-6a. Reaction conditions are the same as shown in Figure 2. Temperature (°C), ○: 25, ●: 50, □: 75.